Biologics are expressed from the hosts of bacterial, yeast, animal cells, and continuous cell lines. As biological products are produced from a cell substrate, it is inevitable that residual host cell DNA is present in the final products. It have potential risks of immunogenicity, oncogenicity, and infectivity due to the tumor or virus-related genes in passage cell lines. Residual DNA has a potential high risk to safety and is considered a critical quality attribute. Current agencies including WHO, EU, and the FDA limited the accepted amounts of residual DNA (less than 10 ng or 100 pg/dose). Among the methods of detecting residual DNA, qPCR is considered to be the most practical for residual DNA quantitation due to its sensitivity, accuracy, precision, and time-saving.
Since rDNA could come from a wide variety of hosts, rDNA may exist in different sizes and physical forms, different limits of DNA content and size may be acceptable to the regulatory agencies if they are supported by scientific evidences and robust risk assessment. The level of host cell DNA residuals can serve as an indicator of product consistency and quality control. Detecting residual DNA ensures consistency between production batches and provides a reference for product quality evaluation.
To analyze host cell DNA concentration and size distribution, DNA fractions are first extracted from the sample to ensure detection without inhibitory compounds from the sample matrix. Magnetic bead separation is the most commonly used technology for this purpose, enabling automated extraction processes that increase throughput and precision.
To determine the extraction efficiency of residual host cell DNA, a known amount of target DNA (the 'Spike') is added to the sample before extraction. By comparing the measured DNA amount after extraction with the spiked amount, the extraction efficiency can be calculated. This efficiency value is then used to determine the residual host cell DNA in the sample. A reduction in extraction efficiency indicates the presence of inhibitory compounds co-extracted with the nucleic acid.
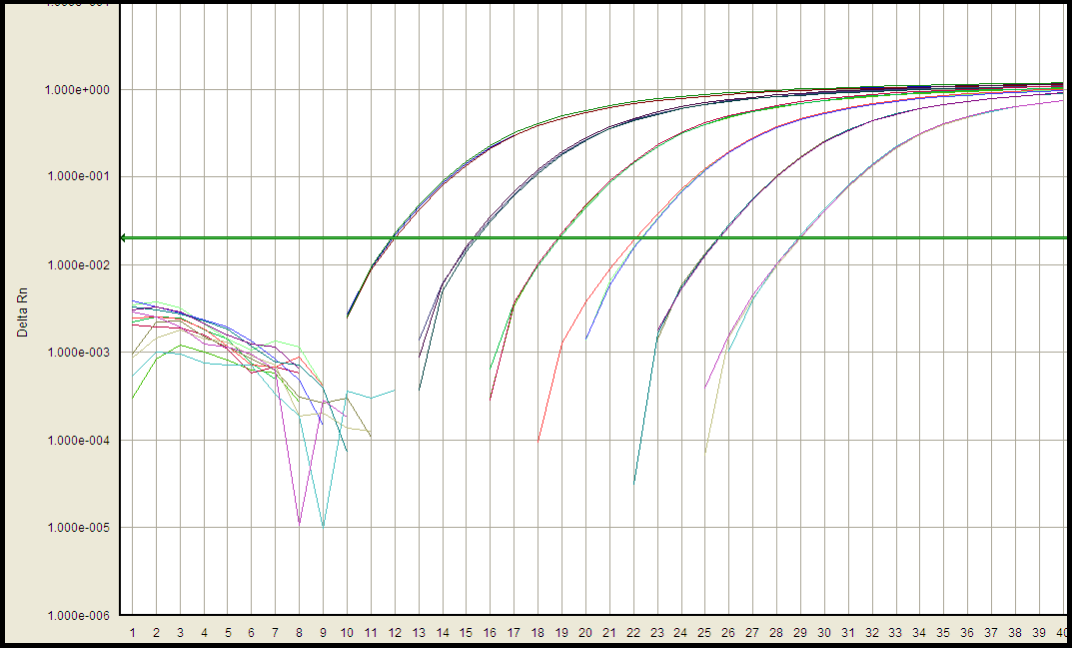
Developing a highly sensitive and specific detection method for host cell DNA using real-time PCR requires careful primer design and PCR reaction optimization. The specificity and sensitivity of real time PCR are influenced by annealing temperature, time, and the concentrations of cations and primers in the reaction buffer. Targeting a gene present in multiple copies can increase assay sensitivity.
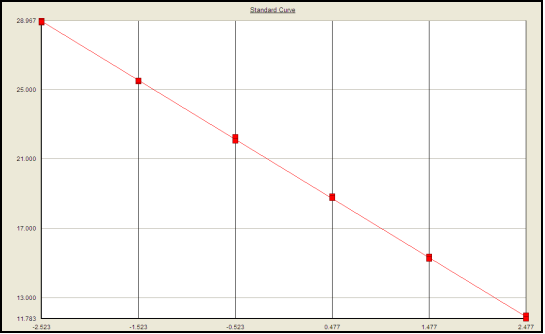
The linearity and range of the assay methods were evaluated by calculating the regression line of the standard curve. Accuracy was assessed by analyzing the recovery percentage of spiked DNA standards at three different concentrations. Intra-assay and inter-assay precision were evaluated via the %CV of all QCs at each level. Sensitivity was determined by checking the LOD and LLOQ. Specificity was assessed by analyzing DNA samples or a pool of matrix DNA extracted from the relevant species/strain. Robustness was evaluated by assessing the stability of assay performance under different storage conditions.
The validation scope of the residual host cell DNA assay includes:
Accuracy
Precision
Specificity
Limit of quantitation (LOQ)
Linearity
Range
Robustness
A fully validated assay allows the use of commercially available products or specifically designed kits targeting the host DNA sequence, making it applicable for testing among biological products.
Case Study—Residual Host Cell DNA Quantitation
CHO HCD Validation performance-
Standard curve: Amplification efficiency 96.6%, Correlation coefficient 0.999